Check-Out / Reservation System for Inventory Items
Inventory application now supports the ability to check-out or reserve containers for current and future use. This will allow users to better plan and track the use of specific inventory items. Other supporting features that expand functionality include the ability to add comments to indicate use, compatibility with Worklists (which was a feature we released last month), search based on checked out status, notifications about inventory availability, warning / error messages for forbidden actions, and audit log of check-in/check-out activity. For more details read this help article on checking out and reserving samples. Contact support@benchling.com if you want to have this feature turned on for your team’s use.
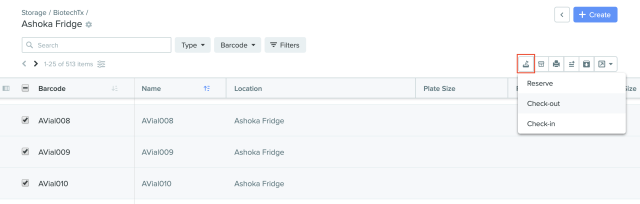
1) Selections for reservation, check-out, and check-in of samples
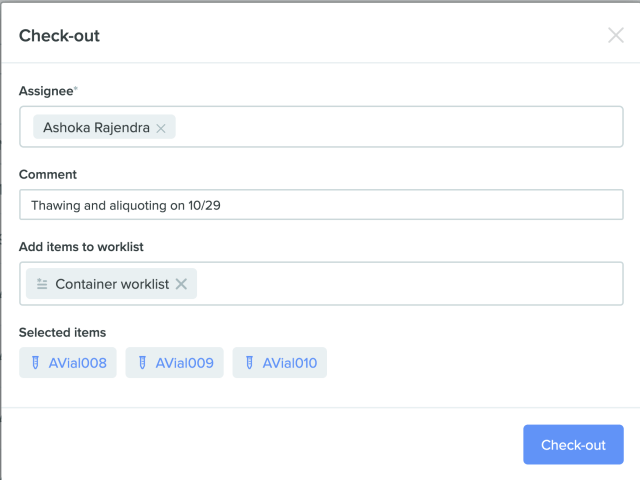
2) Check-out window
Worklist Integration with Bulk Assembly
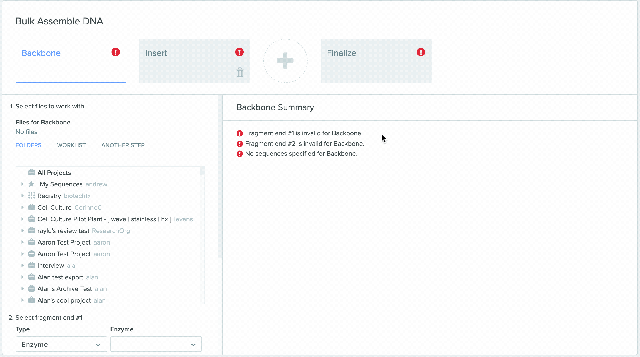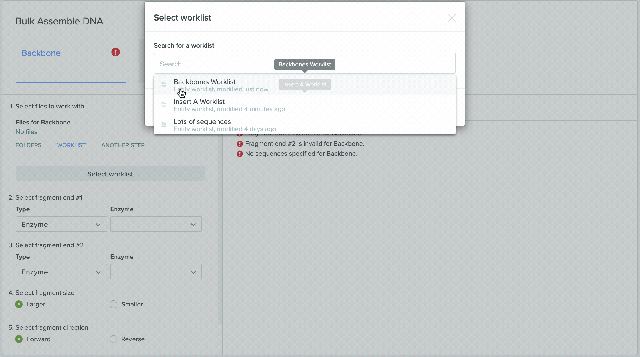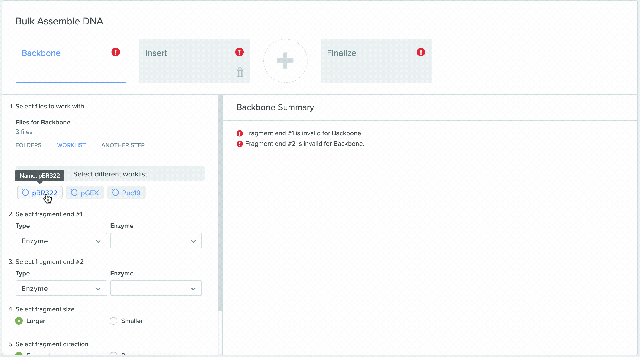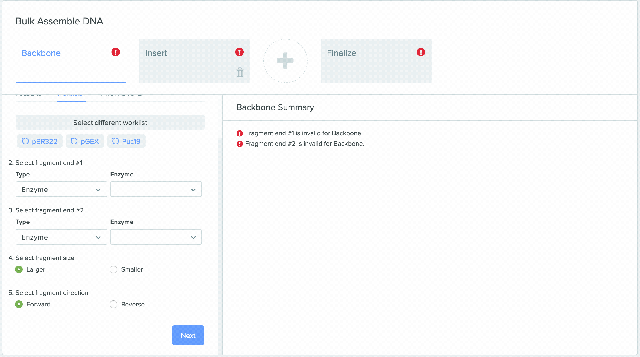
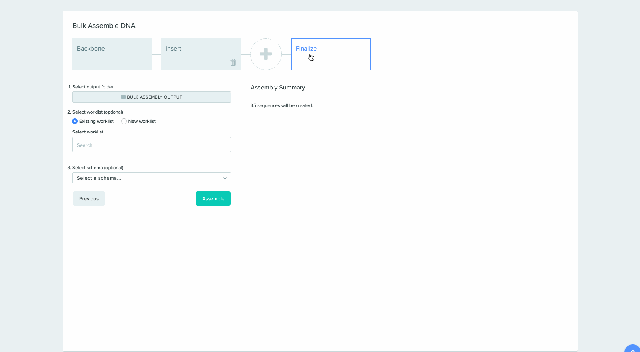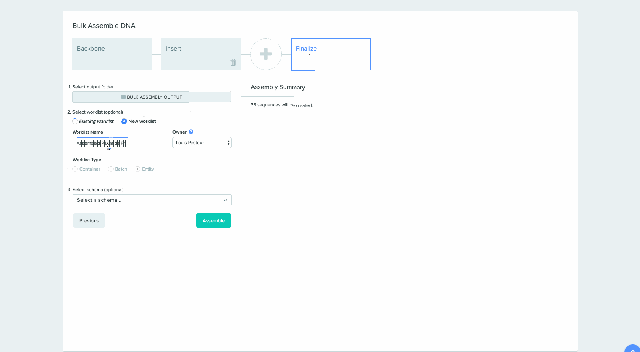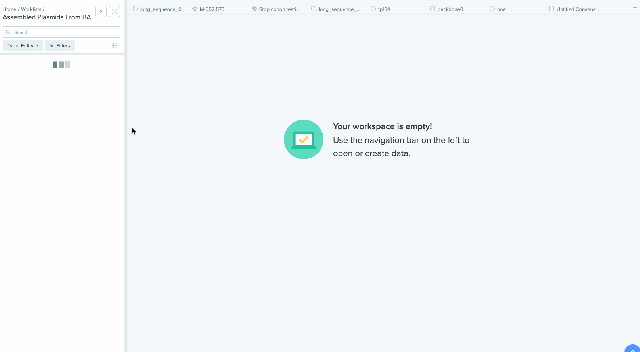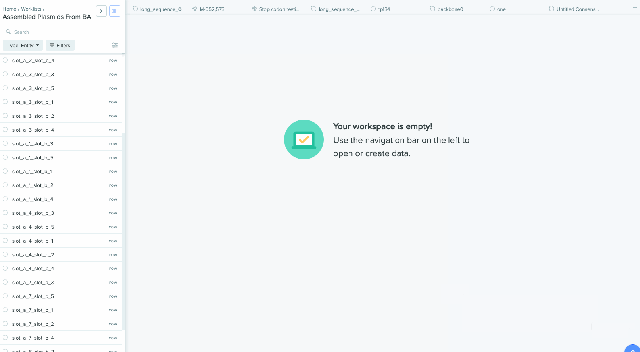
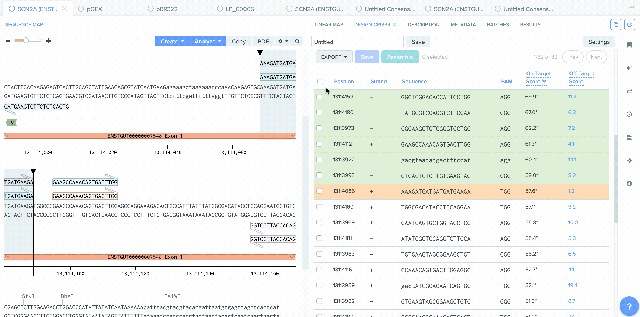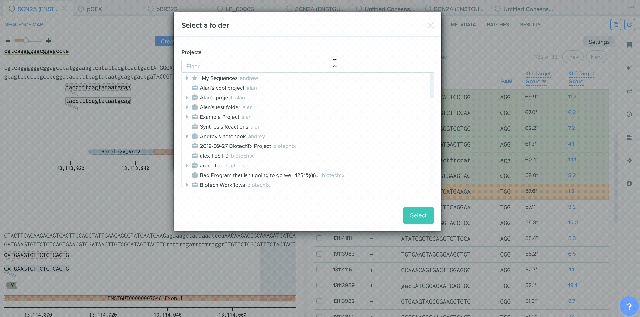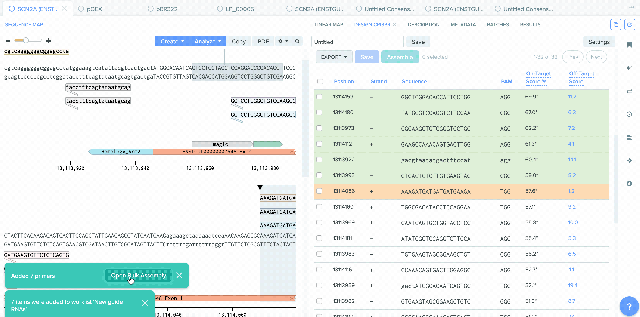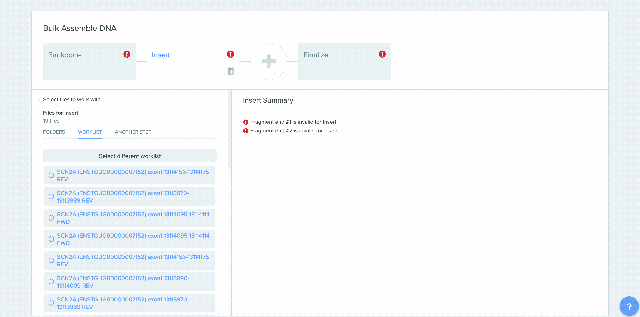
Our Bulk Assembly tool now supports Worklist feature. Users can aggregate all of their desired input sequences into Worklists, and then use these Worklists as input sources into Bulk Assembly. In addition, users can now optionally save the output sequences of their Bulk Assembly into another Worklist. This streamlines the process of selecting sequences from multiple project folders and allows better organization and tracking of all output sequences from a given Bulk Assembly run. This feature is available to all users of the Bulk Assembly Tool. Learn more about Bulk Assembly here.
1) Using Worklist as an input in Bulk Assembly
2) Saving sequences from Bulk Assembly into a Worklist
CRISPR Guide Design Integration with Bulk Assembly
A number of improvements were made to the process of saving CRISPR guides created in Benchling and the integration between CRISPR guide design, Bulk Assembly, and Worklists. sgRNA oligo entities can now be used as input for the Bulk Assembly tool. Also, CRISPR sgRNA created on Benchling or imported from external sources can now be saved into Worklists. These improvements better facilitate the process of assembling guide RNAs into plasmids for expression. This feature is available to all users of the Bulk Assembly Tool. Read up more about designing guide RNAs on Benchling here.
Other Molecular Biology Enhancements
-
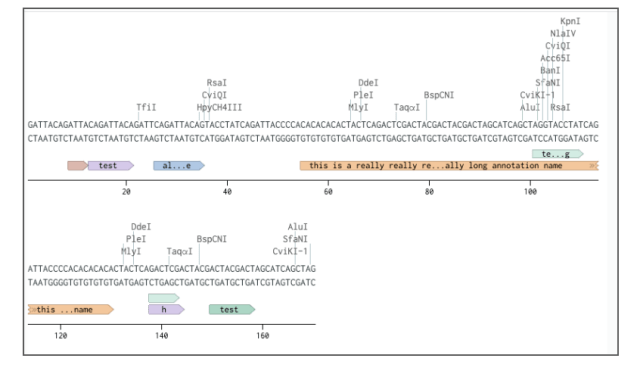
Long annotations assigned to nucleotide and amino acid sequences will now be displayed with truncation in the Molecular Biology application.
-
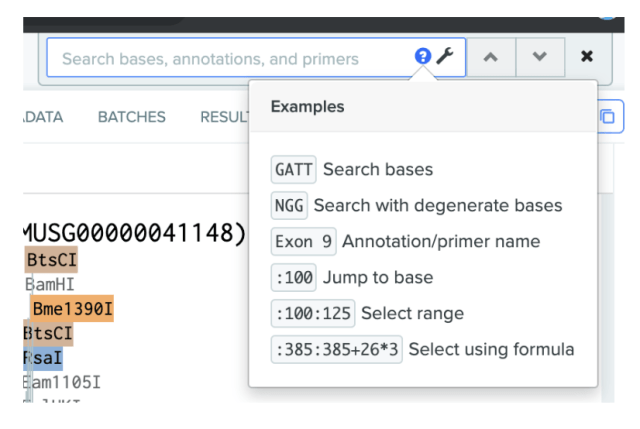
Help text with examples has been added to DNA search to help you use our powerful search function more effectively.
Notebook Enhancements
-
User interface for attachments used with Benchling Sync has been updated. A new box displaying “Checked In” or “Checked Out” has been added to the toolbar to make the status of the attachment more apparent. To learn more about Benchling Sync read this help article.

-
Monospaced fonts to display nucleic acid and protein sequences are now supported in the Benchling Notebook. This feature ensures that the representation of every nucleotide or amino acid occupies the same amount of space that makes it easier to compare different sequences visually.
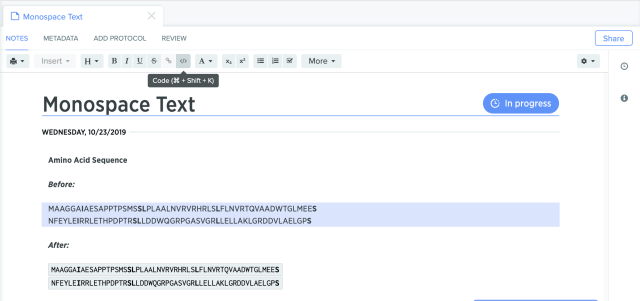
-
Import spreadsheets of entities or batches into a registration table in the Benchling Notebook. Customizable mapping removes the need for manual data manipulation. Registry imports through the registration table in the Notebook can be reviewed and signed-off by reviewers, if required.
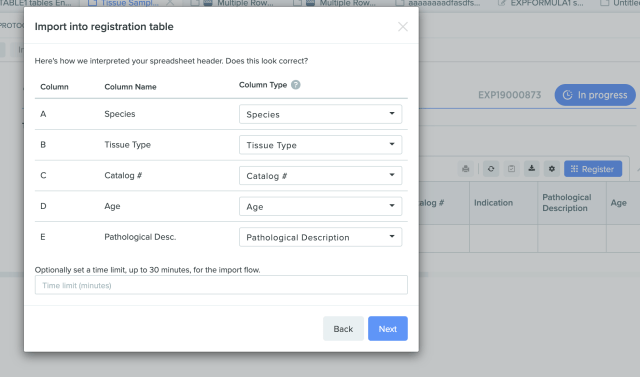
Registry Enhancement
-
Bulk creation of drop down options for registry schemas has been enabled in the Registry. These schemas can be either imported from a spreadsheet or using API calls.
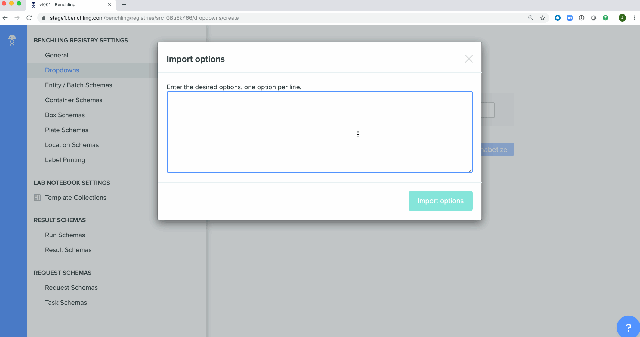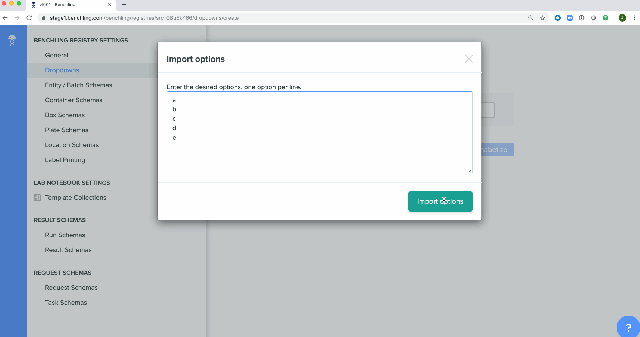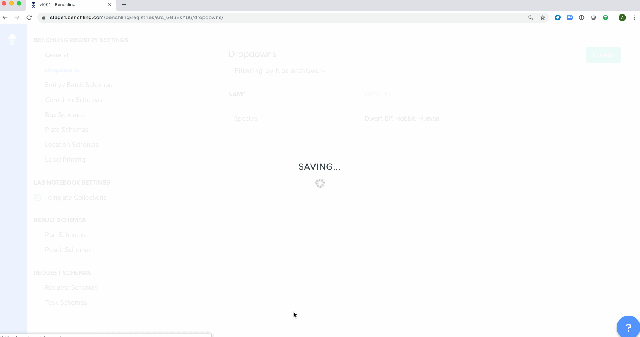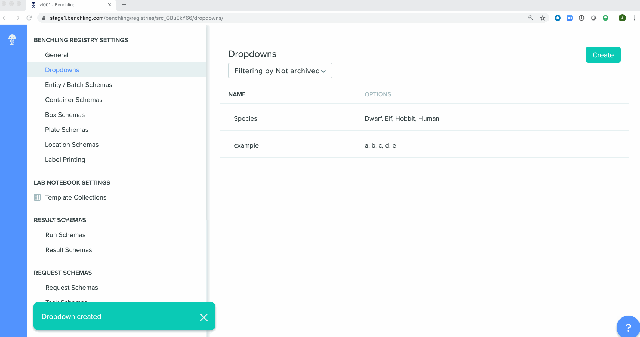
Platform-level Enhancement
-
Email notify relevant team admins by @mentioning teams in your entries. This is in addition to @mentioning individuals that you already love and use on Benchling. Sharing research and collaborating with your colleagues and other cross-functional teams is now easier than ever.
