Monthly Release Notes are published at the end of each month to highlight recent feature additions and updates. Read about our new product features and enhancements for the month of June below.
Insights
Insights is one of our newest applications to the platform. This feature is seamlessly integrated with all Benchling applications, and enables users to query, visualize, and share high quality, structured data that resides on the platform.
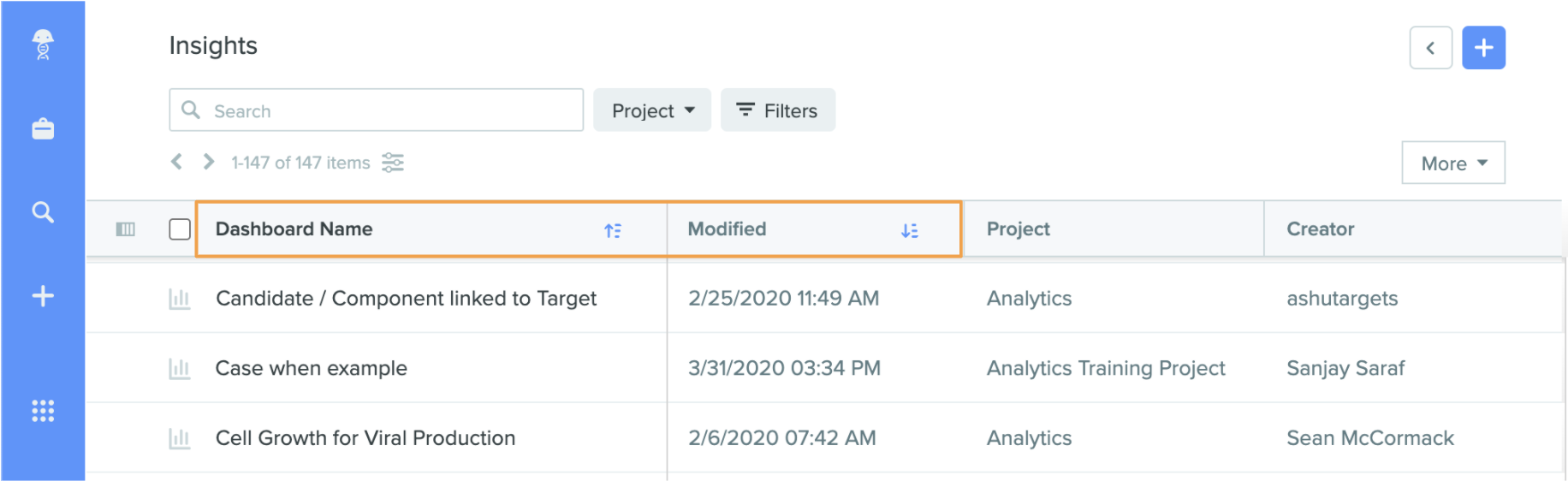
Sort Dashboard Listings by Name or Date Modified
In addition to sorting dashboards by applying filters, dashboard lists can now be sorted alphabetically by name or by last modified in the expanded view. Click on the Dashboard Name or Modified headers at the top of the listing to quickly sort dashboards accordingly.
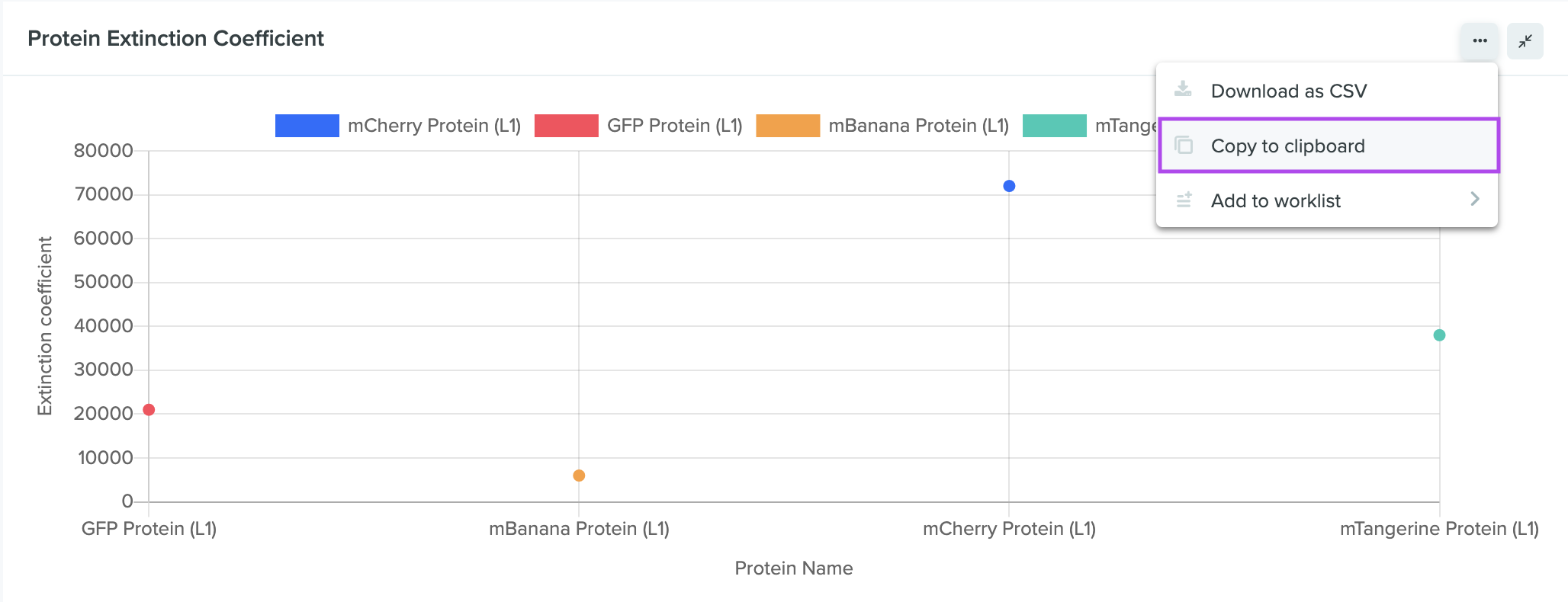
Copy data visualizations directly from the dashboard
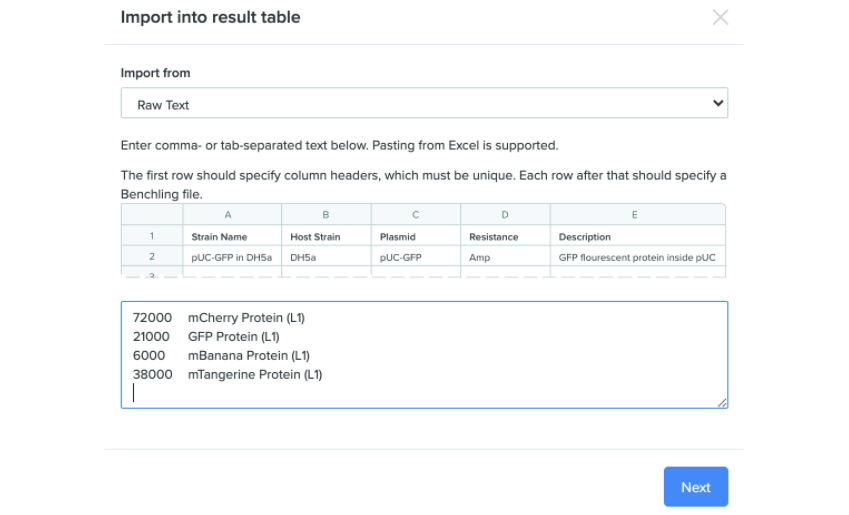
Users can now copy data visualizations directly from the dashboard/query view. Selecting the Copy to clipboard option copies your data visualization as TSV (tab-separated values), allowing for import as raw text once headers are added.
Molecular Biology
Benchling’s Molecular Biology is a cutting edge in silico design tool that supports the latest scientific workflows.
Avoid hairpins in codon optimization
When doing codon optimization, users can now toggle the Avoid Hairpins checkbox to improve translation efficiency. Benchling uses the DNA Chisel sequence optimizer algorithm to identify hairpin RNAs, learn more here.

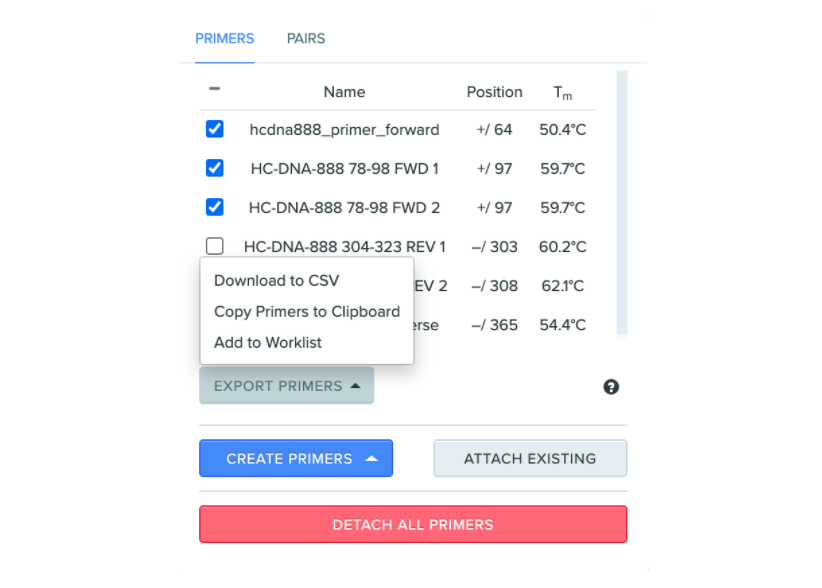
Add selected primers to Worklist from primer panel
Within the primer panel, users can now add selected primers to a Worklist. This feature makes it easier to take bulk actions with primers.
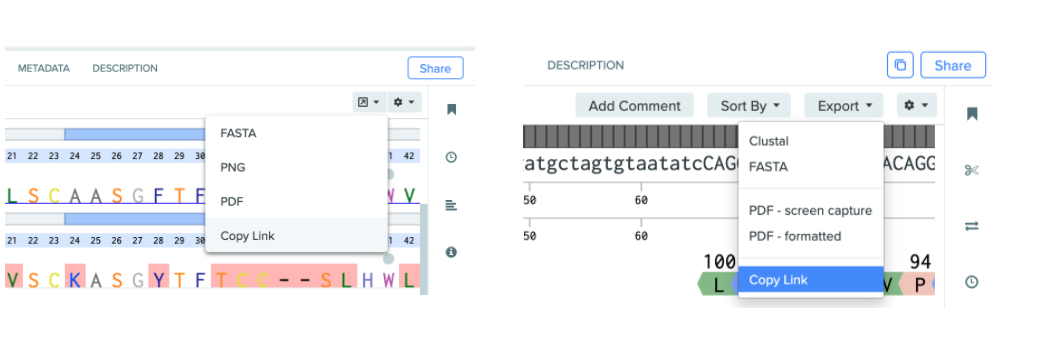
Generate a URL link to any DNA or AA alignment
In the past, alignments were only found on the sequence entity itself, presenting challenges for sharing. Now, users can generate a dedicated URL link to an alignment through the alignment export dropdown. This feature makes it easier to reference a specific alignment in Notebook entries for use by other users.
Developer Platform
Our developer platform is fundamental to centralizing and standardizing all your R&D data. Benchling’s APIs are built to match the flexibility and speed of modern life science R&D.
Update oligos via the API
Adding a PATCH endpoint for oligos allows users to update oligo information, including sequence, name, or metadata fields, using the API. Documentation can be found here.
Return a list of all oligos via the API
Previously, listing oligos separately from DNA sequences was not supported via the API. Adding list endpoint for oligos allows users to return a list of oligos. Documentation can be found here.
Retrieve Blob IDs via the API
Blobs are opaque files (such as PDFs or images) that can be linked to other items in Benchling, such as assay runs or results. Adding GET endpoint for blobs allows users to retrieve the ID of the blob via the API. This will allow users to see all data linked to their assay runs or results via the API. Documentation can be found here.
Unarchive results via the API
Adding unarchive endpoint pulls a list of assay result IDs to be unarchived via the API. Documentation can be found here.
Remove projects from storage items using PATCH endpoints
This feature allows projects to be removed when updating containers, boxes, or plates via the API by setting the projectId to null. Documentation can be found here.
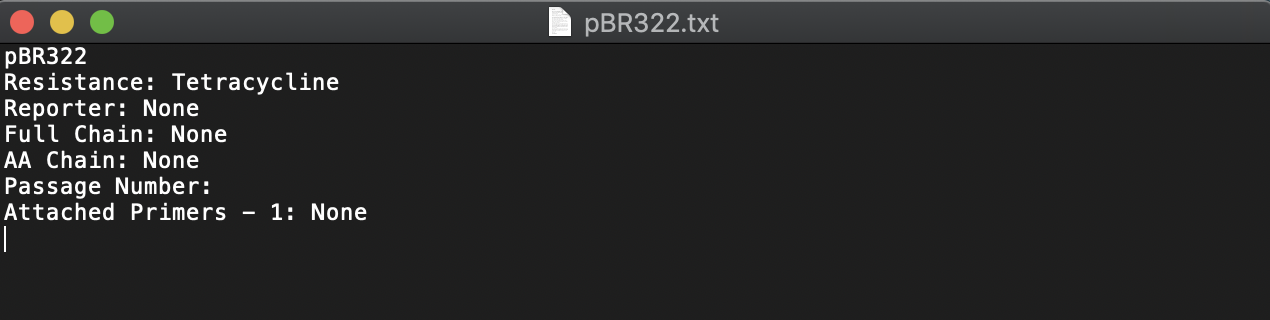
Project Exports Include Sequence Metadata
In the past, exporting a project converted any DNA sequences and AA sequences held within to .gb and .fasta files, respectively; however, metadata such as custom tags and schema fields were not included in the export. This update to project exports offers a separate .txt file for each DNA and AA sequence that lists any custom tags or schema fields. Learn more about exporting projects here.
Retrieve a Single Run via the API
The List Runs endpoint retrieves multiple Runs in the API; however, getting a single Run was not available in the past. Learn more about the new endpoint that retrieves a single run by ID here.
Lab Automation
Benchling for Lab Automation enables seamless integration of Benchling to liquid handlers, plate readers, imaging instruments, and more.
Define Final Concentration Values Using ColumnTypes
ColumnTypes map columns in an output file to a Benchling operation. In the past, processing final dilutions of a sample was dependent on two operations: the measured concentration of a sample before dilution (using the INPUT_CONCENTRATION_VALUE columnType) and the final volume of the sample (defined by FINAL_VOLUME_AMOUNT). The new columnType, FINAL_CONCENTRATION_VALUE, can be applied to directly capture these data after dilution.
Transfer Non-Contained Entities
Previously, lab automation allowed the transfer of an existing sample from one container to another. The addition of the columnType INPUT_BIOENTITY_NAME now allows a non-contained entity to be introduced into a container.
Pivot the Format of Output Files Using a Processing Step
At times, instruments can record results for a single sample in multiple rows, ordered by the quantity of several different measurements (left); however, scientists typically represent their results in a pivoted format, with a single row designated to the sample (right). Pivoting re-orients a dataset by specifying a column whose contents can be turned into column headers. The addition of a pivot step allows users to perform this transformation and orient their data accordingly for lab automation use.

Filter Input Files Based on Schema Field Values
Lab automation has the ability to filter entities based on a certain criteria. Entities that match these criteria can be passed through further actions downstream, such as moving it to another plate for further analysis. The new FILTER type LookupStep will filter samples based on a sample schema field value. This addition is particularly useful for hit-picking workflows.
Support for Processing Output Files Up to 30K Rows
Processing an output file of a Lab Automation Run containing 30,000 results can be uploaded via the API or the UI without performance issues.
Bug Fixes
The following bugs were fixed in this release, ordered by product area:
Insights
-
Benchling supports table visualization with thousands of chips