Before you begin
Learn how to get started and sign up with Benchling here. Start this worksheet using your free academic account in order to get the most out of this worksheet. DNA sequences can be copied directly into your Benchling account. For notebook entries, you can manually copy & paste the content into a new blank entry.
Content and materials for this module were co-developed with Dr. Philip Leftwich, Biology Lecturer at the University of East Anglia (Norwich, UK)
Overview
The central dogma of molecular biology is an essential learning for any scientist. The complex relationships between DNA, RNA, and proteins also mean there are complexities with storing and analyzing those corresponding sequences for experimental purposes as well. While many earlier modules have covered how to create and visualize DNA sequences (genes, plasmids, and templates), this module will cover how to generate and analyze protein sequences on Benchling.
Prior knowledge
You should have a fundamental understanding of the central dogma and how genes can encode for a specific, functional protein. Additionally, you should have awareness of biochemical properties of proteins such as structure, function, folding, isoelectric point (pI), and so on. For a quick overview of proteins and their properties, check out this video.
Learning outcomes
-
Add or remove translation frames over DNA sequences
-
Mutate or change codons for individual amino acids
-
Create and analyze a protein sequence on Benchling
Worked Example
Benchling is a great tool for designing or storing DNA and protein sequences. Better yet, it can help you easily visualize the relationship between a gene and it’s protein counterpart. In this example, we’ll investigate the human BRCA2 gene and understand how to generate its corresponding protein sequence on Benchling and analyze it.
For eukaryotic genes, external databases will allow you to import the full genomic DNA sequence (including exons and introns) or import only the coding DNA sequence (exons only) that comprise the protein.
Import the BRCA2 gene into a Project of your choice along with the following settings and inputs:
-
Genome: GRCh38 (hg38, Homo Sapiens)
-
Transcript: BRCA2-001
-
Click to enable “Import cDNA only”
-
Import As: BRCA2 Gene H. sapiens
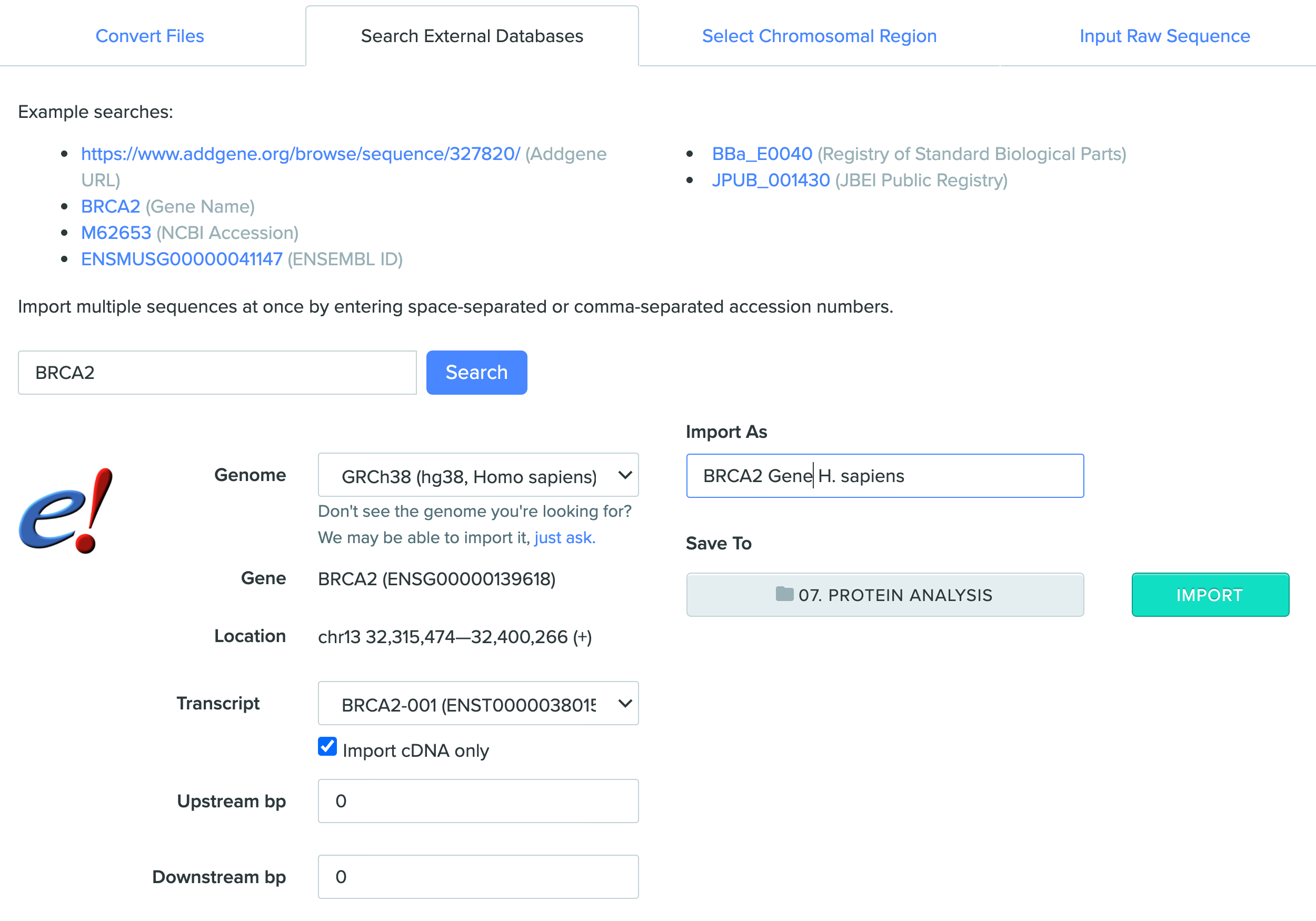
From here, you’ll see a DNA sequence for the BRCA2 gene with an overlay of the protein translation for this gene. Benchling automatically imports any specified protein translations from the file source. Now, let’s explore the gene sequence by doing the following:
-
Toggle on/off different viewing features through the ⚙️ icon like “ORFs” or “Amino Acid Indices” which make it easier to find relevant information.
-
Click on the ORF starting point at the beginning of the BRCA2 gene to highlight the related sequence information.
-
Hover over the DNA sequence and right click to bring up a menu and select “Create Translation”. You’ll notice this creates an additional translation frame alongside the one that was imported with your DNA sequence.
-
Now hover over a specific amino acid in the translation ahd right click to bring up a different menu and select “Change Amino Acid”
Question(s)
1. Within Benchling, you can easily change one codon for one amino acid into a codon that produces a different amino acid. What is the result or the scientific technique called when you’re making singular changes to a protein sequence?
2. With the natural redundancy of multiple codons coding for the same amino acids, can you describe one situation or application where you would want to use different codons for the same amino acid?
Try to answer these question(s) on your own and check the "Solution" at the bottom.
Practice
Let’s do a simple comparison of the BRCA2 gene across different organisms. Use the same settings as before but make sure to specify the following genomes when importing:
-
Rnor_6.0 (Rattus norvegicus)
-
BDGP5 (Drosophila melanogaster)
Browse through the respective sequences and even from a glance, you will notice that the protein sequences across organisms are very different.
Stretch Yourself
Now that you’ve familiarized yourself with looking at genes and understanding the proteins they encode as a DNA sequence, we’ll explore how you can create a corresponding Protein sequence directly on Benchling and analyze them by protein alignments.
Using the BRCA2 (H. Sapiens) DNA sequence, do the following to create a protein sequence:
-
Select the ORF encoding the entire protein and right click and choose “Create AA sequence” -> “Forward”.
-
Specify where you’d like to save the new sequence and then rename the sequence “BRCA2 H. Sapiens Protein” and then hit “Create”.
-
You should now be able to view only the protein sequence without any context of the gene it came from.
-
Check out the tab “Biochemical Properties” to see some computed values that Benchling calculates about this unique protein such as “Molecular Weight”, “Isoelectric Point”, “Extinction Coefficient”
Repeat these steps with the other BRCA2 genes from earlier ((Rattus norvegicus and Drosophila melanogaster) to create protein sequences for them as well. Once you’ve created these sequences, we’ll analyze the homology of all these proteins through a protein alignment.
-
Navigate to your BRCA2 H. Sapiens Protein sequence and on the right-hand toolbar, find the “Alignments” icon and choose “Create New Alignment”.
-
Search for the other protein sequences from other organisms by typing in “BRCA2” and choose the relevant files to do the alignment. You will notice you can also change which sequence is the template but for this example, we’ll use the H. sapiens sequence.
-
Once you’ve loaded all your sequences, choose “Next” and then hit “Create Alignment” to run your protein alignment.
-
Drag the viewer window to look across your protein alignment. You can also change your viewer settings and toggle on/off specific properties and can even export the alignment.
-
The alignment will also summarize statistics such as number of identical sites and pairwise identity between all sequences.
Solution(s)
1. Making singular changes to a protein sequence will yield point mutations for this protein. An example of a research technique that requires making point mutants would be “site-directed mutagenesis”.
2.“Codon optimization” is a common research technique where you interchange various codons that still code for the same amino acid. This is because often different organisms have a bias for which codons are utilized and you will need to optimize the codons for any non-native organism you express a protein in.
Congrats! You've finished the learning module: Protein Analysis.