Purpose
To provide details about the necessary steps to implement the Thermo Chromeleon data connector for use with the Benchling Connect platform.
Introduction
The Thermo Chromeleon data connector is a component used in the context of the Benchling Connect platform to exchange data within the Thermo Chromeleon Chromatography Data System (CDS). Through the use of a Chromeleon ‘Connection’ in Benchling, a user is able to send data from Benchling to populate a sequence within the Chromeleon Console, parse data from Chromeleon to an Allotrope Simple Model (ASM) for liquid chromatography (LC), and make those data available within the Benchling UI in the context of a Run.
The Benchling Connect, Thermo Chromeleon data connector relies on an API-based integration, meaning that it is able to communicate directly with the Chromeleon application via methods provided by the Chromeleon application program interface. The benefit to the user is that once configured the interaction between Benchling and Chromeleon appears more seamless, with few actions that need to be performed by the user during each session.
In order to successfully configure the Thermo Chromeleon data connector for use with the Benchling platform, there are several steps that need to be followed across both the Thermo Chromeleon and Benchling user interfaces.
This guide details the steps to be taken in both applications in order to configure the integration.
Steps within Thermo Chromeleon
Within Chromeleon, a Sequence represents a ‘worklist’ of samples - referred to as Injections - to be processed by the chromatography instrument. The Sequence object and constituent Injections serves as the interface through which the user specifies data acquisition and processing parameters, and retrieves data from previously measured samples.
For configuring the Benchling Connect, Thermo Chromeleon data connector, the principal step that must be performed within the Chromeleon UI is the creation of the directory in which the Benchling Connection will create new sequences and retrieve sequence data from.
Creating Sequence Folder: Chromeleon Console - Data View
Within the Chromeleon UI the Console serves as the starting point from which users access sequences, instruments, methods, reports and other objects represented in the application. data from previous experiments, and create sequences for new experiments.
The Chromeleon CDS relies on a local database for storing Chromeleon objects - referred to as a Data Vault - and displays these objects in a file-tree within the Data View panel of the Console.
From within the Data View panel, a folder should be created to store data to be used in conjunction with the Benchling integration, and a sub-folder specifically for Sequences which will be the location that the Chromeleon data connector will read and write data to. In the example below the ‘_BenchlingConnectTesting’ represents the folder, and ‘Benchling_Sequences’ the sub-folder.
Steps within Benchling
From within Benchling, a user must:
- Enable the Thermo Chromeleon data connector on the tenant [internal admin console]
- Configure a Chromeleon Connection
- Configure Dropdowns and Custom Entities
- Create a Result schema for Chromeleon data
- Configure a Run schema to employ Chromeleon Connection
Enable the Thermo Chromeleon Data Connector
To enable the Chromeleon data connector, as well as any other Benchling Connect data connectors, please reference the Benchling Connect Installation Guide for enabling data connectors via the Internal Admin Console, as well as details related to creating and installing a Gateway and configuring a Connection.
Configure a Chromeleon Connection
To configure a Chromeleon Connection create a new Connection from either the Connections side panel or global Create, and select the Thermo Chromeleon option. The Connection configuration modal prompts the user for (11) fields:
- Connection Name
- Benchling Gateway see previous step
- Sequence Destination - the full path of the Sequence sub-folder
- Vault Root - the full path of the parent folder in the Data Vault containing the sequence sub-folder
- Per User Login Required - 'true' if User Mode is enabled on Chromeleon, 'false' if not
- Username - The username used to login to Chromeleon if a shared account is used for User Mode enabled on Chromeleon
- Password - The password used to login to Chromeleon if a shared account is used for User Mode enabled on Chromeleon
- Report Template (optional) - the name of the report template (from Chromeleon)
- Report Channel (optional) - generating a report template, configure this and report template field. This value is the channel to generate the report for (e.g.
UV_VIS_1) - Benchling Location - location Benchling Files coming off the connection will be created
- Associated Equipment (optional) - associate Benchling equipment with the Connection.
Note that the Sequence Destination and Vault Root are directory paths within the Chromeleon Data Vault and not the Windows Directory. The most reliable way to access this information is to ‘right-click’ the folder within the Data View and select Properties, then select and copy the Location from the Folder Properties modal, and paste into the Connection configuration modal in Benchling.
Configure Dropdowns and Custom Entities
The Benchling Connect, Thermo Chromeleon data connector is able to define (10) properties of a Sequence when creating a new Sequence via the integration. Among these properties is Injection Type. Chromeleon supports an enumerated number of Injection Types and thus it is recommended to define these Injection Types via a Dropdown.
The supported entries for Injection Type are:
- Unknown
- Blank
- CheckStandard
- CalibrationStandard
- Matrix
- Spiked
- Unspiked
Note, that these entries must be defined without spaces (e.g. CheckStandard, not Check Standard).
In order to capture all of the necessary data for generating an Injection via the integration, a ‘Chromeleon Injection’ custom entity should be configured within Benchling.
This custom entity should include:
- Sample Entity - representing the biological entity being assayed
- Injection Type - referencing the Dropdown created in the previous step
- Spike Group - used to associate unspiked (unknown) injections with spiked injections
- Volume - the microliter volume of how much sample should be injected
- Weight - the sample weight to be accounted for in concentration calculations
- Dilution Factor - the correction factor for diluted samples
- Replicate ID - indicator for repeated injections from the same vial
-
Comment - comment field for the injection
Creating Report Template of Chromeleon data
When creating a Report Template from Chromeleon via the Thermo Chromeleon connector to be uploaded into Benchling for a given channel, there a two different ways that this can be configured.
1) Report Template and Report Channel fields are configured via the Connection field definitions (shown above) -- utilized for generic or one time report template creation.
2) Within the Run Schema's Run fields, two fields must be added Report Template and Channel. These can be added to the Run upon processing of your Chromeleon export -- utilized for dynamic report template generation (i.e. differing channels or differing report template names).
To attach the generated report template into your Benchling Entry (Run within the Entry), the Attach additional files Benchling action must be selected in the Run schema Output File configuration
Create Results schema for Chromeleon data
In order to record results returned via the integration a Result schema must be created. This can be done prior to configuration of the Run schema, or within the context of the Run schema Output File configuration.
The Benchling Connect, Thermo Chromeleon data connector extracts data from Chromeleon and formats it according to the Allotrope Simple Model (ASM) for Liquid Chromatography (LC). Note, that the data connector is currently only able to extract data from Chromeleon that was collected using the UV/VIS detector channel.
Details about the ASM LC schema can be found here: https://github.com/Benchling-Open-Source/allotropy/blob/main/src/allotropy/allotrope/schemas/.../liquid-chromatography.schema.json
The data connector then converts the LC-ASM to (3) .csv files: an injection file in which each row represents an injection; a peak file in which each row represents a peak, aggregated across all injections; and a datacube file in which each row represents a chromatogram data point, aggregated by injection.
The injection file can contain the following columns (if available within the data):
- device identifier
- model number [device type (system module such as pump or detector)]
- file name
- UNC path
- software name
- software version
- ASM converter name
- ASM converter version
- column serial number
- analyst
- submitter
- column part number
- device type
- detection type
- reference bandwidth setting (nm)
- reference wavelength (nm)
- detector wavelength setting (nm)
- detector bandwidth setting (nm)
- detector offset
- detector sampling rate
- sample identifier
- sample name
- sample description
- location identifier
- injection
- injection id
- injection time
- injection volume setting
Any CustomField set on the injection will automatically be populated in the INJECTIONS output file.
The peak file can contain the following columns (if available) in addition to the columns within the injection file:
- device identifier
- model number [device type (system module such as pump or detector)]
- file name
- UNC path
- software name
- software version
- ASM converter name
- ASM converter version
- column serial number
- device type -- instrument device type
- detection type
- reference bandwidth setting (nm)
- reference wavelength (nm)
- detector wavelength setting (nm)
- detector bandwidth setting (nm)
- sample identifier
- sample description
- location identifier
- injection identifier
- injection
- injection time
- injection volume setting (μL)
- injection name
- injection position
- injection status
- injection type
- data cube label
- peak name
- peak identifier
- retention time
- peak start
- peak end
- peak height
- peak area
- peak index
- retention time (s)
- relative peak height
- relative peak area
- peak width baseline
- peak width half height
- peak width percentage
- asymmetry factor
- statistical skew
- Capacity factor
- chromatographic peak resolution
- number of theoretical plates by peak width at half height
- peak analyte amount
- chromatographic peak resolution
Any CustomField set on the peak will automatically be populated in the PEAK output file.
In most cases, the peak file will contain the most relevant information.
The example below shows a Results schema capturing: Sample Name, Injection ID, Injection Time, Peak Number, Retention Time, Peak Height, and Peak Area from a peak file.
The datacube file can contain the following columns (if available within the data):
- column part number
- device type
- detection type
- reference bandwidth setting (nm)
- reference wavelength (nm)
- detector wavelength setting (nm)
- detector bandwidth setting (nm)
- detector offset
- detector sampling rate
- sample identifier
- sample name
- sample description
- location identifier
- injection
- injection id
- injection time
- injection position
- injection status
- injection volume setting
- datacube label
- retention time (s)
- absorbance (mAU)
Any CustomField set on the peak will automatically be populated in the DATACUBE output file.
Configure Run schema to employ Chromeleon Connection
The final step is to configure the Run schema to use the Chromeleon integration. This includes:
- Specifying the data connector type
- Defining the Run Fields
- Configuring Input File Processor
- Configuring Output File Processor
Specifying Data Connector Type and Run Fields
When configuring the Run schema for use with the Chromeleon data connector, select Thermo Chromeleon as the Connection Schema. The Run schemas are associated with a type of data connector, rather than a specific Connection, so that Runs can be performed using any Connection of that particular type.
Next, Run fields for Sequence Name, Comment, and Injections should be included. Sequence Name is required as this is the name that will be used for the Sequence created within the Chromeleon UI. Injections is also required and should be designated as a ‘Multi-Select’ so multiple injection entities can be used in the Run. Additionally, Injections should be limited to the ‘Chromeleon Injection’ custom entity created in the previous step. Other Run fields may be added if necessary.
An example of the first portion of the Run schema is shown below.
Configuring the Input File Processor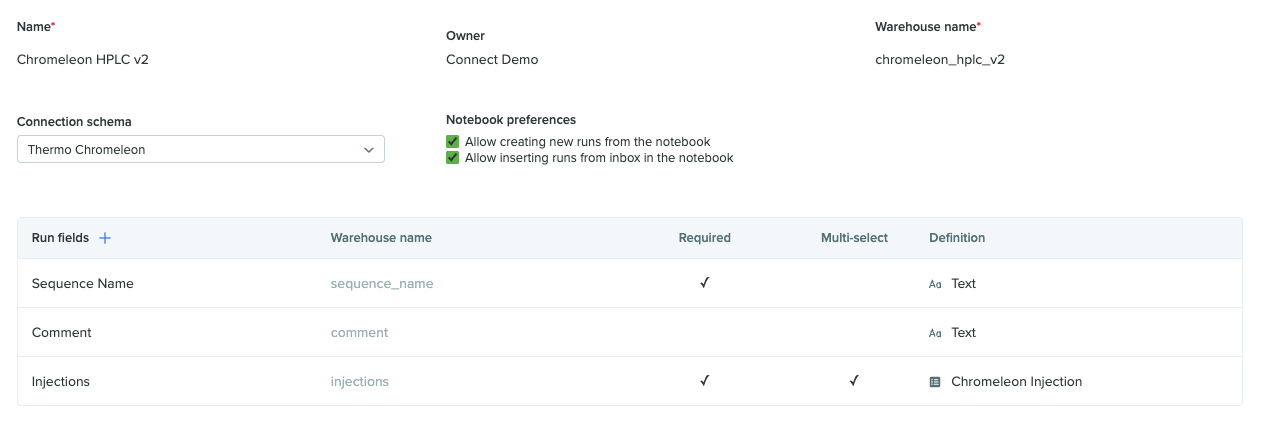
In order to send information from Benchling to Chromeleon, the Run requires an Input File with the requisite information for defining the Sequence. The Input File must have these 10 columns:
- sequenceName
- sequenceComment
- name
- injectionType
- spikeGroup
- volume
- weight
- dilutionFactor
- replicateId
- comment
The sequenceName and sequenceComment are taken from the Sequence Name and Comment Run fields, while the remaining 8 fields should be drawn from LookUps starting from the Injections entry.
As the entries for sequenceName and sequenceComment must be repeated for each injection, ensure that the ‘Consolidate results into single row’ option is selected for these LookUp steps.
Configuring the Output File Processor
The configuration of the Output File Processor is similar to that of other Runs. The Record Results should be specified as the ‘Benchling action’, and the ‘Result schema’ should be set as the schema created in the earlier step.
The column mapping may be manually if the desired columns are known. Alternatively, it is possible to load sample data from an existing Connection to use for column mapping. Additionally, if the Result schema has not been previously defined, it may be done so from this point as well.
As the incoming .csv files have been formatted by the Chromeleon data connector, no formatting transforms should be necessary. However, Custom Transforms or Arithmetic transforms may be employed if additional calculations are required.
An example of the Output File configuration is shown below.
Thermo Fisher Chromeleon Data Connector Requirements
| Item | Specification |
| Chromeleon CDS | Chromeleon 7.2.10* or 7.3.1* |
| Connectivity Type | Chromeleon SDK Specification - Benchling uses the SDK for the “Out-of-process application” |
| (Optional) Chromeleon User Account | Chromeleon user account is required to access Data Servers and Data Vaults when Chromeleon User Mode is enabled |
| Operating System | 64-bit Windows 10 or greater |
| Memory | 8 GB RAM or greater (per Thermo Chromeleon recommendation) |
| Processor | Intel Core i7 or higher (per Thermo Chromeleon recommendation) |
| .NET | .NET Framework >= 4.8 |
| Gateway |
Benchling Windows Gateway installed on PC. The machine hosting the gateway must have connectivity to Chromeleon Data Servers and Data Vaults |
* This is the version of Chromeleon with which the data connector has been developed and tested. The data connector may work with other versions of Chromeleon, but that must be verified by the user.
Revision History
2025/07
Update Connection configuration fields
- Global fields
- Benchling Location (required)
- Associated Equipment (optional)
- Chromeleon Report Parameters
- Report Template (optional)
- Report Channel (optional)
Add section for Report Template generation via Benchling Chromeleon connector
2025/03
Update System Requirements with .NET Framework >= 4.8
Updates to expected output columns for injection and peak data files
Add description for datacube data file csv output with expected output columns
2024/09
Updates to include Chromeleon User Mode support, additional data capture from custom report values
2023/09
Initial Version